Python
Drop Columns
# Drop some useless columns
to_drop = ['state','area_code','phone_number','churned']
churn_feat_space = churn_df.drop(to_drop, axis=1)查看数据的unbalance
# check the propotion of y = 1
print(y.sum() / y.shape * 100)Yes/No转换成True/False
# yes and no have to be converted to boolean values
yes_no_cols = ["intl_plan","voice_mail_plan"]
churn_feat_space[yes_no_cols] = churn_feat_space[yes_no_cols] == 'yes'Gender转换成1/0
X_train['sex'] = (X_train.sex == 'M').astype(int)Encoding Method
get_dummies()函数
多个categorical feature在一列
年份的处理
Train_Test Split
Standardization/Normalization
Classification Problem
Cross Validation
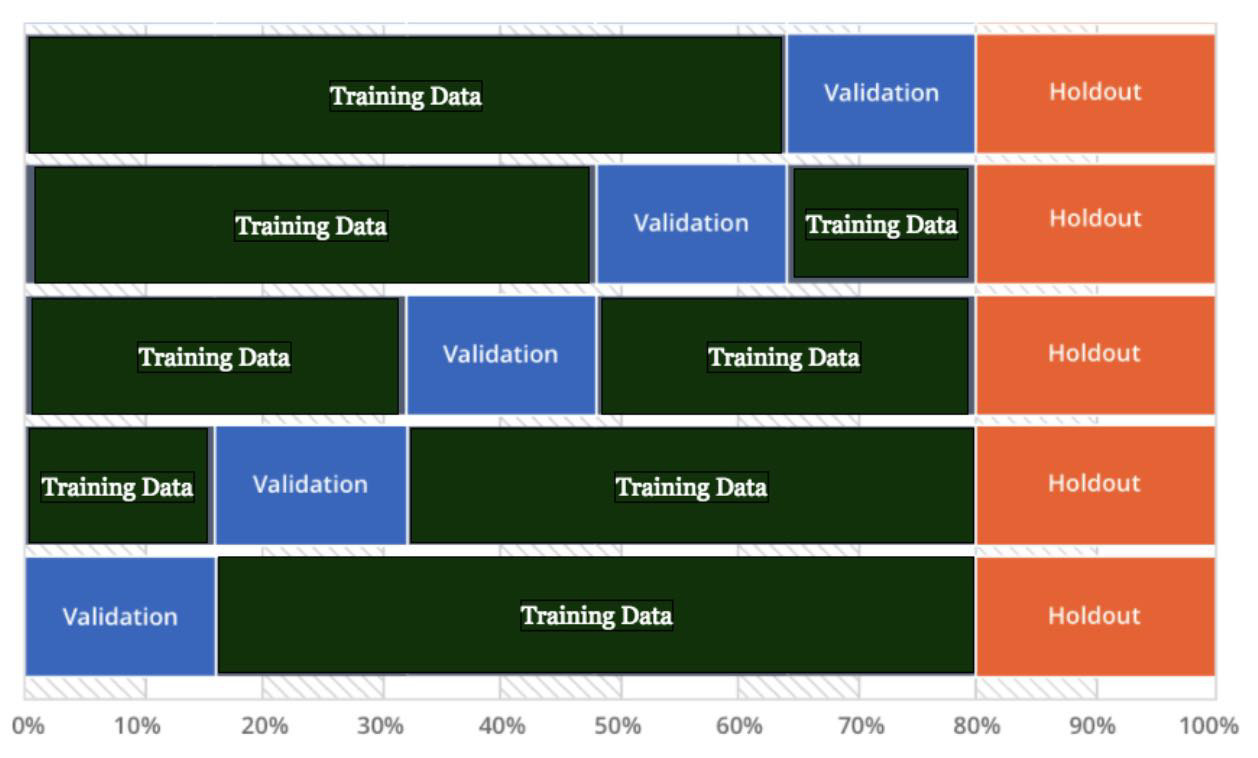
Hyperparameter Tuning
GridSearchCV
Logistic Regression Hyperparameter
KNN
Random Forest
Optimize F1 Score on LR
Optimize F1 Score on RF
Model Evaluation
Confusion Matrix
ROC
AUC
Feature Selection
L1
L2
Random Forest Feature Importance
Regression Problem
Linear Regression
带Polynomial feature的LR
Non-Linear Model: Random Forest
Model Evaluation, score, mean squared error
Lasso, Linear
Ridge, poly
Feature Importance with Random Forest
Unsupervised Learning
K-means clustering
Last updated